Cadmium »
PDB 1gyn-1jre »
1j5l »
Cadmium in PDB 1j5l: uc(Nmr) Structure of the Isolated BETA_C Domain of Lobster Metallothionein-1
Cadmium Binding Sites:
The binding sites of Cadmium atom in the uc(Nmr) Structure of the Isolated BETA_C Domain of Lobster Metallothionein-1
(pdb code 1j5l). This binding sites where shown within
5.0 Angstroms radius around Cadmium atom.
In total 3 binding sites of Cadmium where determined in the uc(Nmr) Structure of the Isolated BETA_C Domain of Lobster Metallothionein-1, PDB code: 1j5l:
Jump to Cadmium binding site number: 1; 2; 3;
In total 3 binding sites of Cadmium where determined in the uc(Nmr) Structure of the Isolated BETA_C Domain of Lobster Metallothionein-1, PDB code: 1j5l:
Jump to Cadmium binding site number: 1; 2; 3;
Cadmium binding site 1 out of 3 in 1j5l
Go back to
Cadmium binding site 1 out
of 3 in the uc(Nmr) Structure of the Isolated BETA_C Domain of Lobster Metallothionein-1
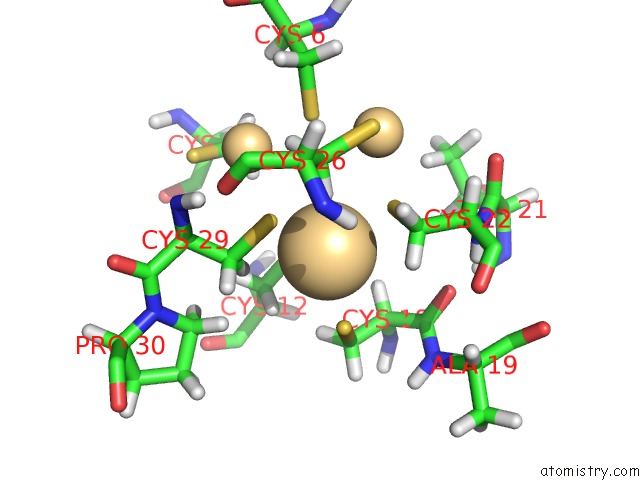
Mono view
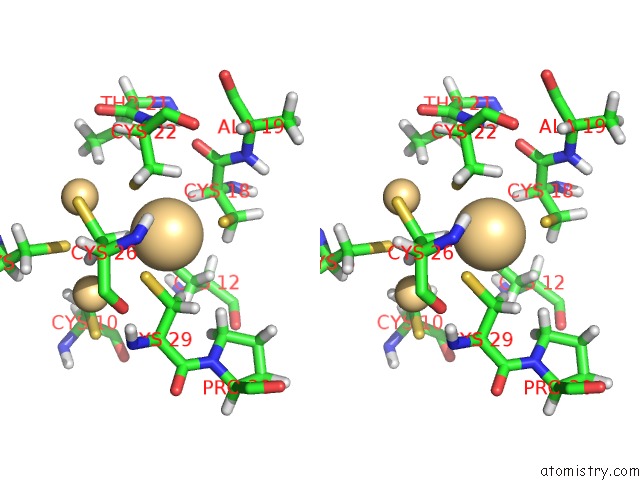
Stereo pair view

Mono view

Stereo pair view
A full contact list of Cadmium with other atoms in the Cd binding
site number 1 of uc(Nmr) Structure of the Isolated BETA_C Domain of Lobster Metallothionein-1 within 5.0Å range:
|
Cadmium binding site 2 out of 3 in 1j5l
Go back to
Cadmium binding site 2 out
of 3 in the uc(Nmr) Structure of the Isolated BETA_C Domain of Lobster Metallothionein-1
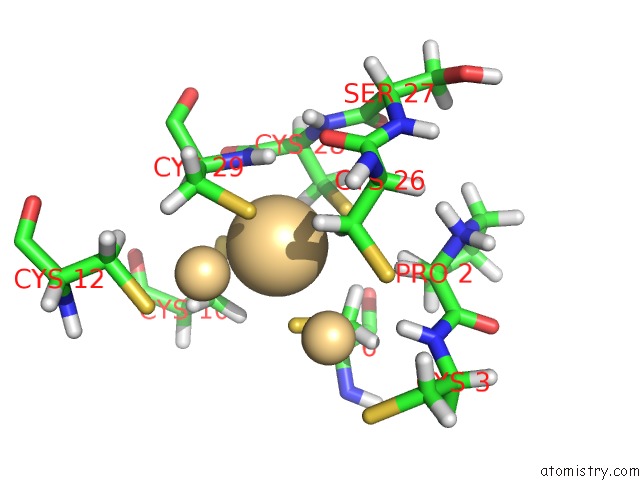
Mono view
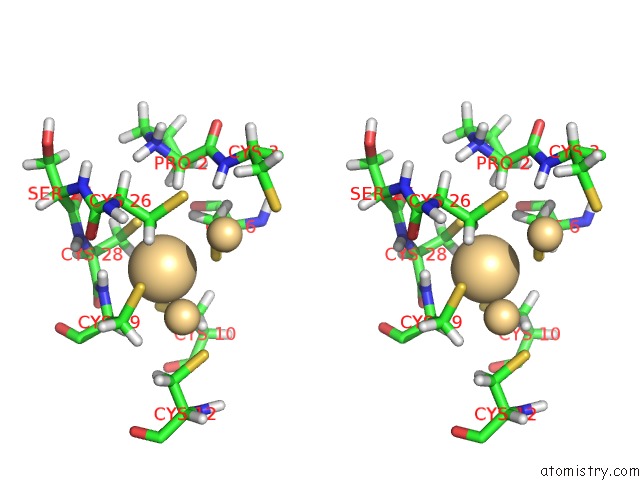
Stereo pair view

Mono view

Stereo pair view
A full contact list of Cadmium with other atoms in the Cd binding
site number 2 of uc(Nmr) Structure of the Isolated BETA_C Domain of Lobster Metallothionein-1 within 5.0Å range:
|
Cadmium binding site 3 out of 3 in 1j5l
Go back to
Cadmium binding site 3 out
of 3 in the uc(Nmr) Structure of the Isolated BETA_C Domain of Lobster Metallothionein-1
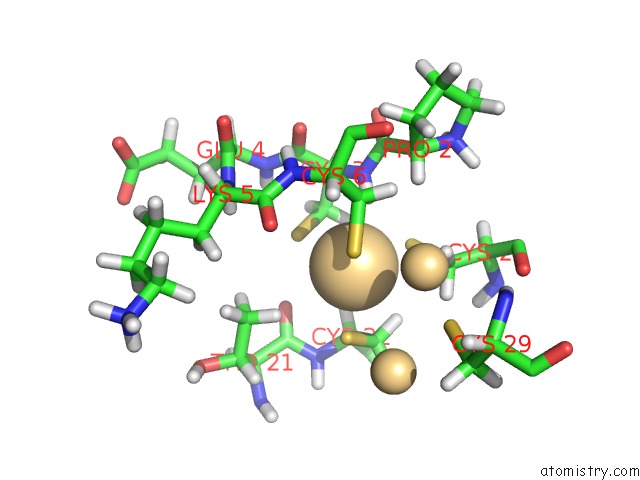
Mono view
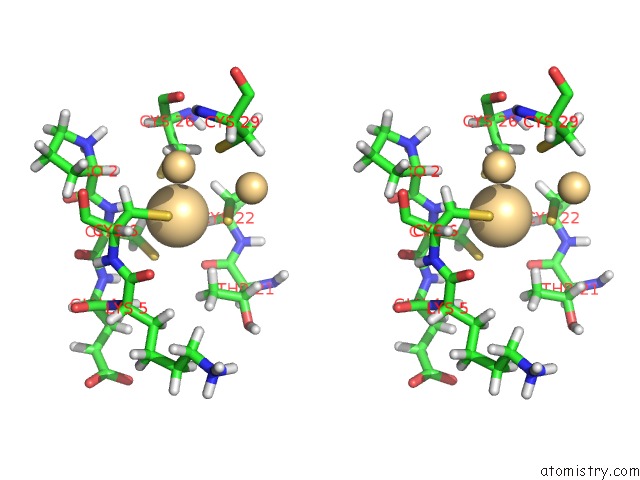
Stereo pair view

Mono view

Stereo pair view
A full contact list of Cadmium with other atoms in the Cd binding
site number 3 of uc(Nmr) Structure of the Isolated BETA_C Domain of Lobster Metallothionein-1 within 5.0Å range:
|
Reference:
A.Munoz,
F.H.Forsterling,
C.F.Shaw Iii,
D.H.Petering.
Structure of the (113)Cd(3)Beta Domains From Homarus Americanus Metallothionein-1: Hydrogen Bonding and Solvent Accessibility of Sulfur Atoms J.Biol.Inorg.Chem. V. 7 713 2002.
ISSN: ISSN 0949-8257
PubMed: 12203008
DOI: 10.1007/S00775-002-0345-3
Page generated: Thu Jul 10 10:52:25 2025
ISSN: ISSN 0949-8257
PubMed: 12203008
DOI: 10.1007/S00775-002-0345-3
Last articles
Cl in 2XJNCl in 2XJP
Cl in 2XGL
Cl in 2XJM
Cl in 2XI4
Cl in 2XIY
Cl in 2XHR
Cl in 2XIW
Cl in 2XIK
Cl in 2XEW